The Power of Folding
This is where the power of Folding@home proved itself. Using six months of computer time on Folding@home, Pande was able to run thousands of simulations, and then analyse the speed of the dozen or so proteins that successfully folded. Some were slower or faster than others, but Gruebele explains that "by knowing how many trajectories he [Pande] had run, and what fraction of them had folded, he was able to extract an accurate folding time for the simulation."Gruebele adds the disclaimer that this doesn’t mean that the time he got from the simulation "was an accurate measurement, but at least he was actually able to get a time. That’s what he was able to do that people had not been able to do before – predict that this protein will take X microseconds plus or minus."
Meanwhile, Gruebele and his team at Illinois performed their experimental measurements, and the two teams then compared their results, which turned out to be surprisingly close. "To be honest," says Gruebele, "it was even closer than I would have ever thought." Folding@home had proved that it could accurately simulate protein folding, with results that could be verified by traditional experimental testing. A paper called ‘Absolute comparison of simulated and experimental protein-folding dynamics’ detailed the tests, and was published in ‘Nature’ in 2002.
Gruebele and Pande have stayed in touch since then, and Gruebele says he’s now in talks with Stanford about using Folding@home to perform computational simulations of how proteins fold when placed under pressure. Similar collaborations with other experimental researchers have also been performed since then, while others are in the process of being verified. There’s a full page of 63 results papers on Stanford’s website, but Pande also says that some interesting discoveries have also been made that haven’t been published.
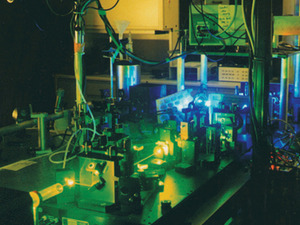 The image to the right shows the experimental setup used for the BBAW experiment in Martin Gruebele’s lab in 2002. Gruebele explains that "the yellow/green laser light in the foreground is pumping an infrared laser (invisible), which is then nonlinearly doubled (blue background glow) and tripled (invisible ultraviolet) to be sent into the protein sample. The emission of the protein then reports on the folding status every14 nanoseconds."
The image to the right shows the experimental setup used for the BBAW experiment in Martin Gruebele’s lab in 2002. Gruebele explains that "the yellow/green laser light in the foreground is pumping an infrared laser (invisible), which is then nonlinearly doubled (blue background glow) and tripled (invisible ultraviolet) to be sent into the protein sample. The emission of the protein then reports on the folding status every14 nanoseconds."Pande describes a recent test carried out by Brian Dyer from the Los Alamos National Laboratory in 2008 that was similar to the Gruebele blind test. The findings haven’t been published, but Pande says that they have been publicly discussed, and that Dyer "showed that many of the predictions that we made are bearing out to be verified in his most recent experiments". There are also some potentially very exciting results from collaborations with experimental labs that haven’t been publicly disclosed.
In December 2008, ‘The Journal of Chemical Physics’ published a paper from Stanford’s Folding@home team called ‘Simulating oligomerization at experimental concentrations and long timescales: A Markov state model approach’.
The paper made some specific predictions about a new type of therapy for Alzheimer’s, and the predictions are now in the process of being tested in experimental labs to verify them. Although the results haven’t been written up yet, Pande is clearly very excited about the findings so far, and says he’s "very optimistic about the outcome". However, as a professional scientist, he said that he couldn’t go into further detail until the work was published.

MSI MPG Velox 100R Chassis Review
October 14 2021 | 15:04









Want to comment? Please log in.